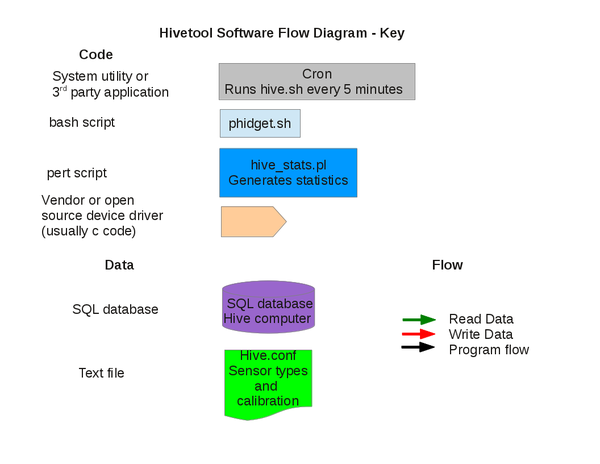
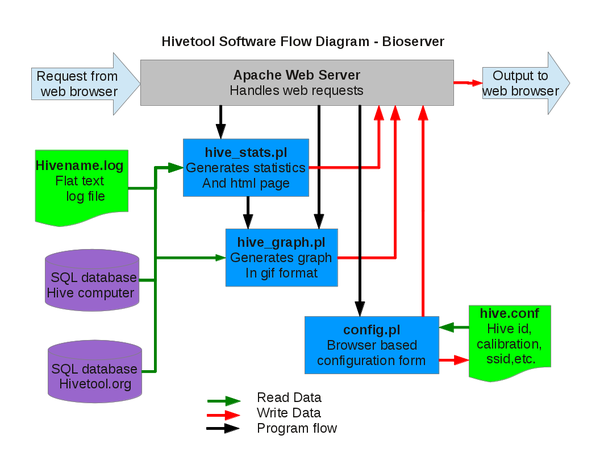
Software: Overview
Software is continuously developed (this page is always out of date). For detailed installation instructions, see How to load hivetool on the Pi. This project uses Free and Open Source Software (FOSS). The operating system is Linux although everything should run under Microsoft Windows. The code is available at GitHub.
Hivetool can be used as a:
- Data logger that provides data acquisition and storage.
- Bioserver that displays, streams, analyzes and visualizes the data in addition to data acquisition and storage.
Both of these options can be run with or without access to the internet. The bioserver requires installation and configuration of additional software: a web server (usually Apache), the Perl module GD::Graph, and perhaps a media server such as Icecast http://www.icecast.org/ or FFserver http://www.ffmpeg.org/ffserver.html to record and/or stream audio and video.
Linux Distributions
Linux distros that have been tested are:
- Debian Wheezy (Pi)
- Lubuntu (lightweight Ubuntu)
- Slackware 13.0
Data Logger
Scheduling
Every 5 minutes cron kicks off the bash script hive.sh that reads the sensors and logs the data.
Initialization
Starting with Hivetool ver 0.5, the text file hive.conf is read first to determine which sensors are used and to retrieve their calibration parameters.
Reading the Sensors
Hive Weight
Several different scales are supported. For tips on scales that use serial communication see Scale Communication. Scales based on the HX711(see Frameless Scale or the AD7193 (Phidget Bridge) Analog to Digital Converter are supported on the Pi.
Temperature and Humidity Sensors
tempered reads the RDing TEMPerHUM USB thermometer/hygrometer. Source code is at github.com/edorfaus/TEMPered Detailed instructions for installing TEMPered on the Pi.
Weather
cURL is used to pull the local weather conditions from Weather Underground in xml format and write it to a temporary file, /tmp/wx.html. The data is parsed using grep and regular expressions.
Logging the Data
After hive.sh reads the sensors and gets the weather, the data is appended to a flat text log file hive.log and written in xml format to the temporary file /tmp/hive.xml by xml.sh. cURL is used to send the xml file to a hosted web server where a perl script extracts the xml encoded data and inserts a row into the database. Starting with version 0,5, the data is also stored in a local SQL database by sql.sh.
Bioserver
Just as a mail server serves up email and a web server dishes out web pages, a biological data server, or bioserver, serves biological data that it has monitored, analyzed and visualized.
Visualizing the Data
GD::Graph
The Perl module GD::Graph is used to plot the data. The graphs and data are displayed with a web server, usually Apache. More detailed installation instructions are on the Forums.
Filters
Filters are used to remove "noise" and distortion from the data.
Displaying the Data
Apache Web Server
When a request from a web browser comes in, the webserver kicks off hive_stats.pl that queries the database for current, minimum, maximum, and average data values and generates the html page. Embedded in the html page is a image link to hive_graph.pl that queries the database for the detailed data and returns the data in tabular form for download or generates and returns the graph as a gif. hive_graph.pl can be called as a stand alone program to embed a graph in a web page on another site.